 |
 |
||||||
   |
|
|
VariantSEQr®
|
VariantSEQr™ SystemApplied Biosystems has discontinued selling re-sequencing probes through the VariantSEQr and MitoSEQr product lines. IntroductionState-of-the-art capillary fluorescent sequencing is the most accurate technique for identifing DNA sequence variations among individuals in a population. The VariantSEQr™ primer sequences and experimental protocol provided by Applied Biosystems provides effective means to PCR-amplify and sequence a large fraction of human genes. Primer DesignThe target sequences of the Resequencing Amplicons (RSA) have been designed to provide complete coverage of promoter regions, intron/exon junctions and all exons. This is based on the merged annotation from the NCBI and Celera Genomics. Primer design is driven by the primer3 program (Whiltehead Institute for Biomedical Research) and additional algorithms developed by Applied Biosystems. Candidate primers are screened to avoid known SNP sites and then checked for genomic redundancy to avoid secondary amplification products. Primer pair performance is predicted by rules derived from experimental testing of over 200,000 amplicons in earlier studies by Celera Genomics.
How It Works 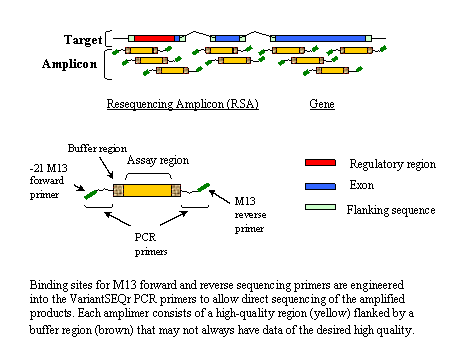
Each successful resequencing amplicon (RSA) is given a unique name (e.g. RSA001488497) by Applied Biosystems and is assigned an NCBI ProbeDB accession number (e.g. Pr001034461.1) upon submission to the database (the .1 suffix is a version number that may later be incremented if the sequence changes). For each gene, a resequencing set (RSS) is defined to provide a minimal tiling set of amplicons for efficient sampling of the gene. Additional RSAs not part of the minimal set are also provided as possible alternatives. ResultsThe initial VariantSEQr data release includes 429,931 RSAs total, 289,049 of which are partitioned into 15,525 RSSs (genes). An average gene may be covered by approximately 19 RSAs. Overall, about 90% of the intended target bases were covered (buffer regions are not included in coverage statistics). Resequencing ProtocolThe VariantSEQr System Protocol consists of 5 steps: (1) PCR amplification of human genomic DNA, (2) purification of PCR the product, (3) cycle sequencing, (4) electrophoresis on a sequencing instrument, and (5) data analysis with the SeqScape® software. Note that the protocol conditions may require modification based on available reagents and equipment. ResourcesApplied Biosystems Links » VariantSEQr™ System Overview » VariantSEQr™ Experimental Protocol (.pdf) For additional information, please write to: VariantSEQr@appliedbiosystems.com ProbeSample QueriesReferenceLin,X., Ni,J., Zhang,Z., Rydland,M.A., Sanders,R.D., Merkulov,G.V., Hoover,J.R., Shen,P., Sorenson, J., Edwards,N., Glanowski,S., Forbes,C., Gire,H., Ma,P., Fang,R., Heiner,C., Pham,L.Z., Lakdawalla,A.A., Li,P.W. and Spier,E.G. A comprehensive set of primers designed for the PCR and sequencing of human genes. Unpublished (2005). DisclaimerMention of specific products or vendors on this website does not constitute an endorsement by the U.S. government. |
Questions or Comments?
E-mail the NCBI Service Desk