 |
 |
||||||
   |
|
|
SSO probes
|
Sequence-Specific Oligonucleotide (SSO) ProbesIntroductionSequence-Specific Oligonucleotide (SSO) probes are used in many important hybridization techniques including DNA microarrays designed for large-scale variation analysis and genotyping of complex DNA samples. Single nucleotide polymorphisms (SNPs) are the most abundant form of genetic variation in the human genome and play a critical but as yet largely uncharacterized role in human disease. Rough estimates suggest that there are 10-30 million SNPs in the human genome, or 1 about every 100–300 bases, on average . Millions of human SNPs have been catalogued and made available in public repositories such as the NCBI dbSNP and International HapMap Project (HapMap). Brief review of SNP discovery and genotyping techniques can be found here. Below is description of SSO hybridization-based SNP genotyping approach used in Affymetrix GeneChip® assay. How It Works SNP-containing regions are amplified from genomic DNA, cleaved, tagged, and hybridized to the probe array under stringent conditions, with washing followed by fluorescent labeling. 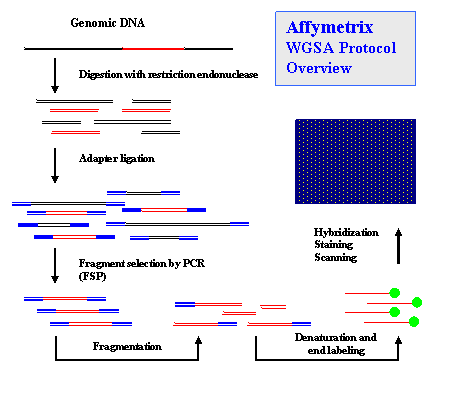 For detailed protocol descriptions, see References and HapMap data protocols. 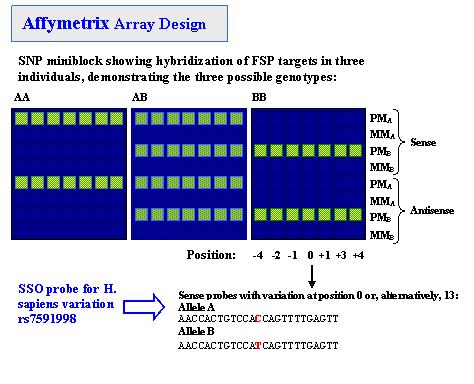 Probes (25-mers) are synthesized as perfect matches (PM) and as one-base mismatches (MM). A perfect match for each of the two SNP alleles (A, B) and one mismatch for each of the two alleles with the nucleotide in question at a given position are referred to as a probe quartet. There are a total of 56 probes for a SNP miniblock: seven probe quartets containing one SNP at seven different positions along both strands of the sequence. It is possible to reduce the number of probes per SNP from 56 to 40 without loss of accuracy. Note: only perfectly matched probes are deposited in the NCBI ProbeDB. In some cases, either sense or antisense probes with the SNP at central, 0th (13th) position are present on the probe report page, and the rest of the probes are omitted. ProbeSample QueriesReferences» Kennedy GC, et al. Large-scale genotyping of complex DNA. Nat Biotechnol. 2003 Oct;21(10):1233-7. Epub 2003 Sep 7. PMID: 12960966 »
Matsuzaki H, et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. » Matsuzaki H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004 Nov;1(2):109-111. PMID: 15782172 ResourcesPubMed query: Note: [MAJR] is a Medical Subject Heading (MeSH) tag for Major Heading. The tag is used to limit the search to articles for which major subjects are represented by terms included in the NLM MeSH database. Terminology and Abbreviations
DisclaimerMention of specific products or vendors on this website does not constitute an endorsement by the U.S. government. |
Questions or Comments?
E-mail the NCBI Service Desk