 |
 |
||||||
   |
|
|
Bead Arrays
|
Bead ArraysIntroductionIn the field of microarrays, planar substrates are most widely used. However, there are several companies developing alternative arraying strategies. Bead arrays are created by either impregnating beads with different concentrations of fluorescent dye, or by some type of barcoding technology. The beads are addressable and used to identify specific binding events that occur on their surface. Illumina (San Diego, CA, USA) has developed a novel bead array technology. A multicore optical 'imaging' fiber is etched such that a bead can fit into the resulting micron-sized etched wells on the tip of the fiber. Different oligonucleotide sequences are attached to each bead, and thousands of beads can be self-assembled on the fiber bundle. A subsequent decoding process is carried out to determine which bead occupies which well. Complementary oligonucleotides present in the sample bind to the beads, and bound oligonucleotides are measured by using a fluorescent label. Illumina cells these arrays in two formats: Sentrix Array Matrix and Sentrix BeadChips (highest density array platform currently available). Illumina participates in International HapMap Project. BeadArray platform (GoldenGate™ protocol) developed by Illumina is used by Broad, Illumina, Sanger, McGill, Beijin, Shanghai/Taipei (more about this protocol). Illustration of the GoldenGate™ genotyping assay process. 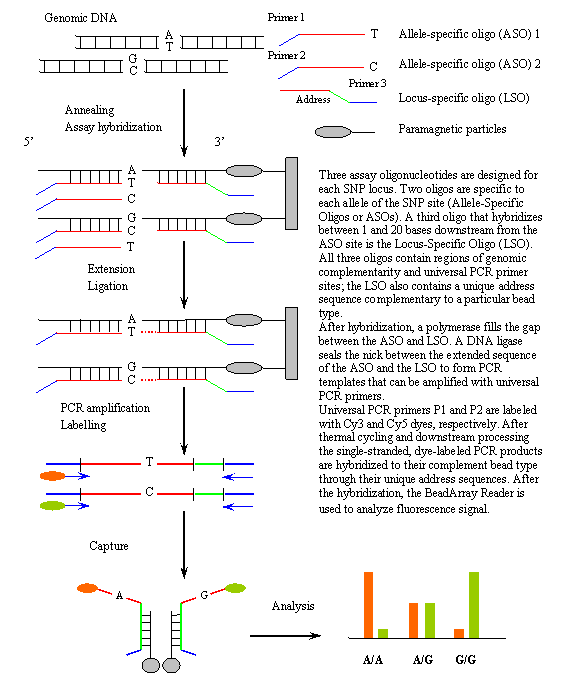
ProbeSample Queries
Resources» More about bead arrays in PubMed |
Questions or Comments?
E-mail the NCBI Service Desk