 |
 |
||||||
   |
|
|
MIP
|
Molecular Inversion Probe AssayIntroductionMolecular Inversion Probe (MIP) Assay is and efficient technology for large-scale Single Nucleotide Polymorphisms (SNPs) analysis. It is used for SNP discovery and genotyping. This technique produces "inverted" probes in which SNPs are introduced into tag sequences that could be analyzed using a universal sequence tag. This technology allows to perform multiplex analysis of more than 10,000 probes in a single tube can be done using standard laboratory equipment. Molecular Inversion Probes (originally called "padlock probes") are single-stranded DNA molecules containing two regions complementary to regions in the target DNA that flank SNP in question. Each probe also contains universal primers' sequences separated by endoribonuclease recognition site and a 20-nt tag sequence. During the assay the probes undergo a unimolecular rearrangement: they are (1) circularized by filling gaps with nucleotides corresponding to the SNP in four separate allele-specific polymerization (A, C, G, and T) and ligation reactions; (2) linearized in enzymatic reaction. As a result they become "inverted". This step is followed by PCR amplification. Further processing of the probes depends on specific assay. How it works 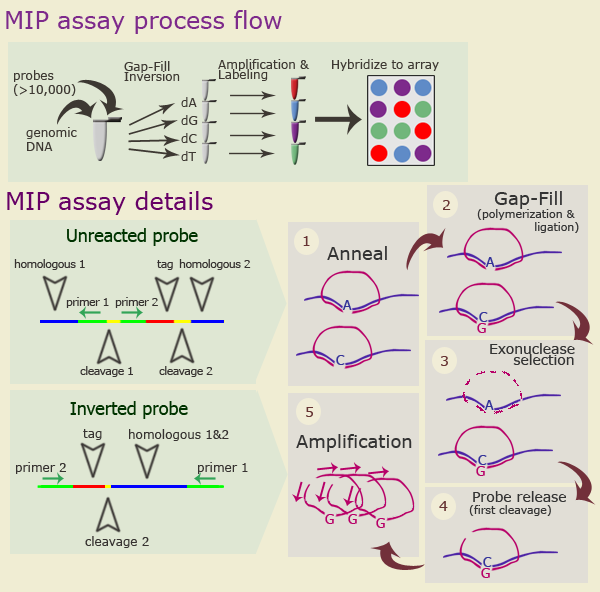
References» Nilsson M et al. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994 Sep 30;265(5181):2085-8. PMID: 7522346 » Syvänen AC. Toward genome-wide SNP genotyping. Nat Genet. 2005 Jun;37 Suppl:S5-10. PMID: 15920530 » Absalan F, Ronaghi M. Molecular inversion probe assay. Methods Mol Biol. 2007;396:315-30. PMID: 18025701 » Hardenbol P et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003 Jun;21(6):673-8. Epub 2003 May 5. PMID: 12730666 |
Questions or Comments?
E-mail the NCBI Service Desk