|
 |
||||||
   |
|
|
STS
|
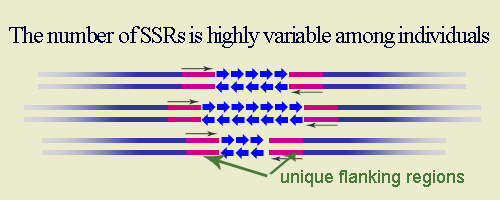
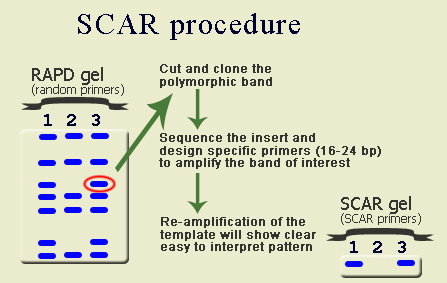
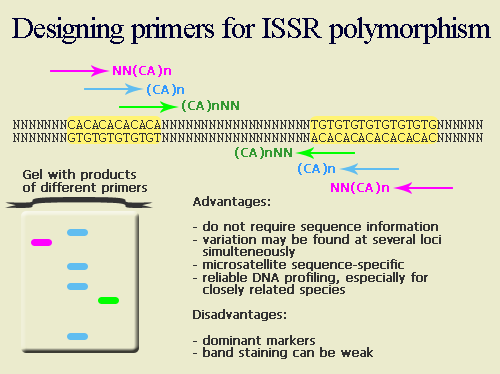
Sequence-Tagged Sites (STS)IntroductionThe Sequence-Tagged Site (STS) is a relatively short, easily PCR-amplified sequence (200 to 500 bp) which can be specifically amplified by PCR and detected in the presence of all other genomic sequences and whose location in the genome is mapped. The STS concept was introduced by Olson et al (1989). In assessing the likely impact of the Polymerase Chain Reaction (PCR) on human genome research, they recognized that single-copy DNA sequences of known map location could serve as markers for genetic and physical mapping of genes along the chromosome. The advantage of STSs over other mapping landmarks is that the means of testing for the presence of a particular STS can be completely described as information in a database: anyone who wishes to make copies of the marker would simply look up the STS in the database, synthesize the specified primers, and run the PCR under specified conditions to amplify the STS from genomic DNA. STS-based PCR produces a simple and reproducible pattern on agarose or polyacrylamide gel. In most cases STS markers are co-dominant, i. e., allow hetorozygotes to be distinguished from the two homozygotes. The DNA sequence of an STS may contain repetitive elements, sequences that appear elsewhere in the genome, but as long as the sequences at both ends of the site are unique and conserved, researches can uniquely identify this portion of genome using tools usually present in any laboratory. Thus, in broad sense STS include such markers as microsatellites (SSRs, STMS or SSRPs), SCARs, CAPs, and ISSRs. Definitions
ProbeSample Queries
References» Olson M et al. A common language for physical mapping of the human genome. Science. 1989 Sep 29;245(4925):1434-5. PMID: 2781285 Resources» The NCBI Electronic PCR (e-PCR) compputational tool » PubMed query: "Sequence Tagged Sites"[Majr] DisclaimerMention of specific products or vendors on this website does not constitute an endorsement by the U.S. government. |
Questions or Comments?
E-mail the NCBI Service Desk