|
Real Time qRT-PCR
|
Real-Time qRT-PCR
Introduction
Real-Time qRT-PCR (Real Time quantitative Reverse
Transcription PCR) is a major development of PCR technology that enables reliable detection and measurement of products generated during each cycle of PCR process. This technique became possible after introduction of an oligonucleotide
probe, which was designed to hybridize within the target sequence. Cleavage of the probe during PCR because of the 5' nuclease activity of Taq polymerase can be used to detect amplification of the target-specific product.
How It Works
 |
The following techniques can be used to monitor degradation of the probe:
- intercalation of double-stranded DNA-binding dyes
- 32P probe labeling
- labeling of the probe with fluorescent dyes
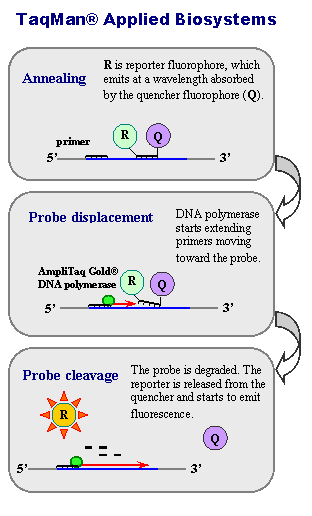
TaqMan
assay (named after Taq DNA polymerase) was one of the earliest methods introduced for real time PCR reaction monitoring and has been widely adopted for both the quantification of mRNAs and for detecting variation. The method exploits the 5' endonuclease activity of Taq DNA polymerase to cleave an oligonucleotide probe during PCR, thereby generating a detectable signal. The probes are fluorescently labeled at their 5' end and are non-extendable at their 3' end by chemical modification. Specificity is conferred at three levels: via two PCR primers and the probe. Applied Biosystems probes also include a minor groove binder for added specificity.
Applications of real time quantitative RT-PCR: relative and absolute quantification of
gene expression, validation
of DNA microarray results,
variation analysis, counting bacterial, viral, or fungal loads, etc.
|
|
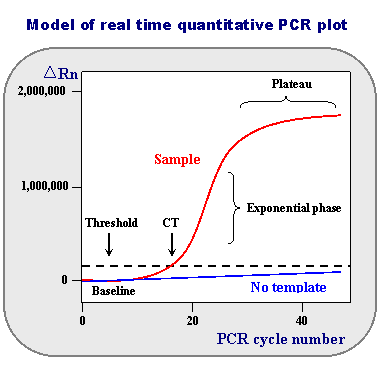
Nomenclature commonly used in real time quantitative RT-PCR:
Baseline is defined as PCR cycles in which a reporter fluorescent signal is accumulating but is beneath the limits of detection of the instrument.
ΔRn is an increment of fluorescent signal at each time point. The ΔRn values are plotted versus the cycle number.
Threshold is an arbitrary level of fluorescence chosen on the basis of the baseline variability. A signal that is detected above the threshold is considered a real signal that can be used to define the threshold cycle (Ct) for a sample. Threshold can be adjusted
for each experiment so that it is in the region of exponential amplification across all plots.
Ct is defined as the fractional PCR cycle number at which the reporter fluorescence is greater than the threshold. The Ct is a basic principle of real time PCR and is an essential component in producing accurate and reproducible data.
|
Probe
Resources
» Heid, CA,Stevens J, Livak, KJ, Williams, PM. 1996 Quantitative Real Time PCR. Genome Res 6:986-994.
PMID: 8908518
» About PCR technology in PubMed
Disclaimer
Mention of specific products or vendors on this website does not constitute
an endorsement by the U.S. government.
|






